Mutations in NUP160 Are Implicated in Steroid-Resistant Nephrotic Syndrome
In the news:
NUP160 genetic mutation linked to steroid-resistant nephrotic syndrome (American Association for the Advancement of Science (AAAS))
NUP160 genetic mutation linked to steroid-resistant nephrotic syndrome (BrightSurf.com)
Patient clinical features:
A 10-year-old girl was admitted to the Department of Nephrology, Shanghai Ruijin Hospital for persistent proteinuria that began at age 7 years old. The proband’s renal specimen revealed focal segmental glomerulosclerosis (FSGS), and IF staining showed nonspecific scattered mesangial deposits of IgA and IgM. The proband progressed to end-stage renal disease (ESRD) and underwent renal transplantation. A 13-year follow-up of the graft kidney showed normal renal function and no post-transplant reoccurrence of nephrotic syndrome. The proband’s pedigree delineated an autosomal recessive family with steroid-resistant nephrotic syndrome (SRNS), which consisted of healthy parents, the proband, two siblings affected with SRNS, two siblings who died of unknown causes in early childhood and one healthy sibling.
Patient genetic variant:
Based on genomic sequence data, the possibility that the proband carried mutations in genes already known to be associated with SNRS was excluded. Using bioinformatics-based analysis of minor allele frequency, mutation type, clinical characteristics and web-based pathogenic prediction the candidate gene list was narrowed to six genes. We ultimately focused on compound-heterozygous mutations NUP160-R1173X and NUP160-E803K because the proband only displayed renal issues without syndromic extrarenal symptomatology. Both NUP160 variants were absent in 520 control subjects. The R1173X variant had been identified previously in an unrelated Chinese family with nephrotic syndrome. To provide functional evidence to validate the NUP160 variants as pathogenic, we used our Drosophila cardiac nephrocyte experimental model to study the effects of NUP160 gene deficiency in vivo.
Precision Drosophila disease model:
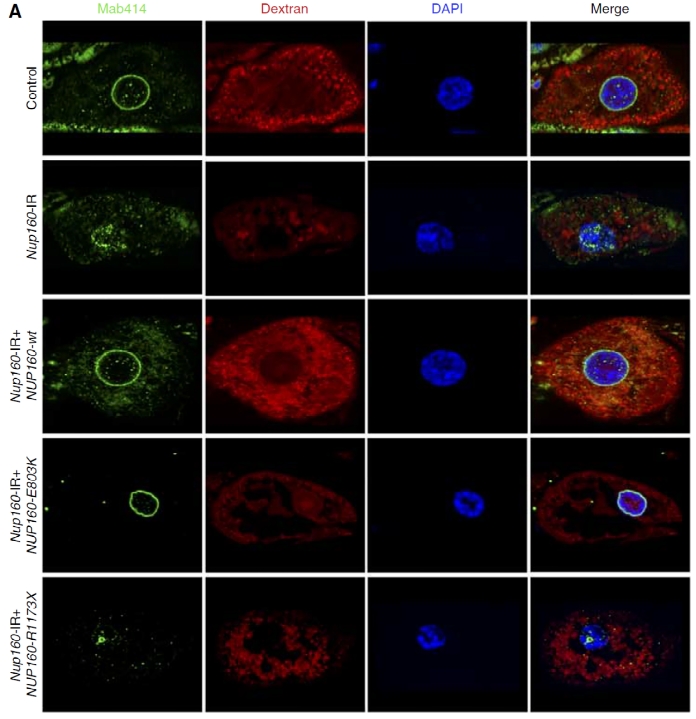
First, we expressed the human NUP160 cDNA transgene (genetic reference) specifically in nephrocytes (fly equivalent of kidney cells) while simultaneously silencing endogenous fly Nup160 expression. Reference human NUP160 expression completely rescued Nup160-IR–induced depletion of adult nephrocytes in flies (Figure 1B). Moreover, the restored nephrocytes were fully functional based on ANF-RFP uptake assay. By contrast, neither the patient-derived NUP160 disease variant allele E803K or R1173X was capable of rescuing the adult nephrocyte phenotype (Figure 1C).
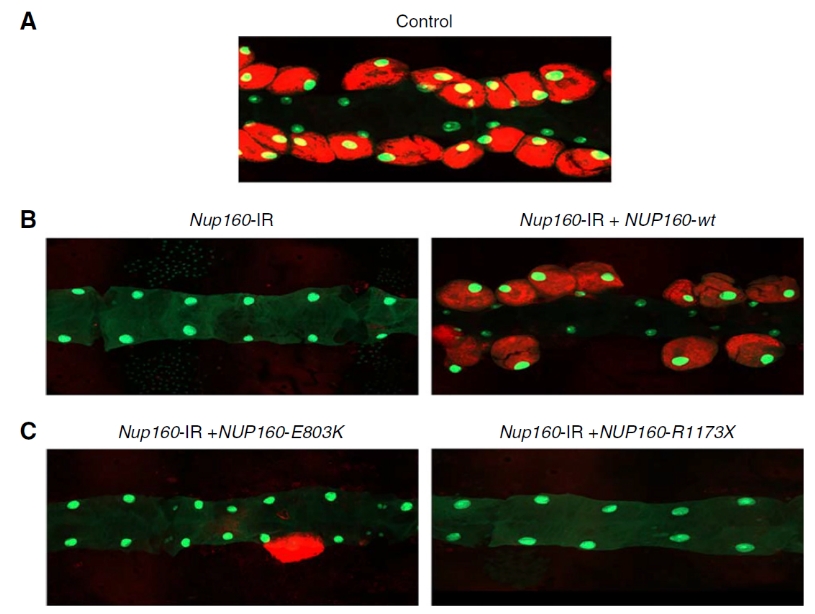
Figure 1. Adult nephrocyte phenotypes induced by Nup160-IR were rescued by reference human NUP160 transgene, but not by NUP160-E803K or NUP160-R1173X. Fluorescence micrograph of nephrocytes from adult flies, 1-day post-emergence. A GFP transgene (green; mostly nuclear) is expressed under the control of a Hand gene enhancer (Hand-GFP) to confirm pericardial nephrocyte cell identity. All flies are transgenic for Hand-GFP. Atrial natriuretic factor (ANF)-RFP fluorescence expression (red) is driven by muscle-specific MEf2-Gal4. ANF-RFP is secreted into the hemolymph (fly equivalent of blood) and its uptake be nephrocytes is measure of their function. (A) Control flies carry the Dot-Gal4 driver but no RNA interference construct. (B, left panel) Nephrocyte-specific Nup160-IR transgene expression to silence endogenous fly Nup160. (B, right panel) Nephrocyte-specific expression of the reference human NUP160 transgene (NUP160-wt). (C, left panel) Nephrocyte-specific expression of disease-variant NUP160-E803K transgene. (C, right panel) Nephrocyte-specific expression of disease-variant NUP160-R1173X transgene.
Further investigation showed that the reference human NUP160 expression was able to completely rescue the phenotype of silenced fly Nup160 in third instar larvae, including the defects in nuclear volume, Dextran uptake, NPC localization and nuclear Lamin morphology (Figure 2). In contrast, neither patient-derived NUP160 disease-variant allele was able to do so. Interestingly, the NUP160-E803K disease-variant transgene did demonstrate rescue of NPC localization and nuclear Lamin morphological phenotypes. These findings suggest the E803K missense allele produces a Nup160 protein that can promote NPC assembly and support nuclear Lamin, and potentially nuclear membrane morphology, however it still results in defective nucleocytoplasmic transport with severe consequences for nephrocyte cellular structure and function. Conversely, the NUP160-R1173X transgene was unable to rescue any of the phenotype, indicating the truncated R1173X allele is essentially null.