Chemical Genetics
Chemical genetics involves the discovery, development and use of chemical probes for interrogation of biological processes and for translational discoveries. In a manner analogous to classic forward mutagenesis screens, the Hong lab has conducted an unbiased, high-throughput chemical screen for small molecules that specifically modulate early embryonic development in zebrafish, and has carried out the follow-up task of identifying the pharmacological targets of a number of developmental modulators.
The zebrafish embryo is a small aquatic vertebrate that allows us to dose the animals in a high throughput fashion while using minimal resources. And their external fertilization and development, and transparent body negate the need for surgeries to extract embryos and allows for investigation of the entire body plan without using complicated staining protocols, thereby increasing throughput. Below is a short timelapse video of the first 24hours of development for a zebrafish embryo.
Zebrafish Develop Rapidly
In addition to the screening and identification of novel molecules we also work with 1) computational methodologies to aid the screening process, 2) the target deconvolution, and 3) the understanding of ligand-target interactions.
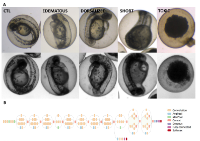
1) The screening methodology employed currently requires manual scoring of phenotypes. This type of methodology represents variability and error, caused by the scorer. Each scorer has a bias towards what they are most familiar with, and the experience of each scorer varies widely from undergraduates with a few weeks of zebrafish experience to veterans with over a decade of experience. And finally, the descriptions are typically sparse and qualitative and highly variable. To address this issue we are developing novel systems to do phenotypic screens. These methods include a set of deep neural networks trained to identify specific phenotypes. By incorporating machine vision principles and automation we hope to reduce phenotyping errors and increase reliability and throughput.
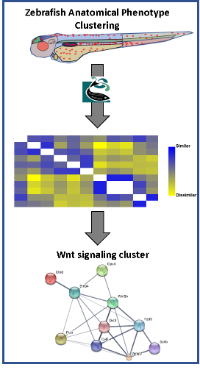 2) The identification of what biomolecule a small molecule interacts with to cause a phenotypic change is referred to as “target deconvolution”. Traditional modes of target deconvolution include affinity based approaches that are highly variable in their effectiveness and are incredibly labor intensive. This step is the transition from teratology to chemical biology, as such is also a step of value generation. To address this we are developing a tool based on classical forward genetics principles, that “like phenotypes will be in shared pathways”. This algorithm utilizes the wealth of mutant and knockdown data that is publically available for zebrafish as qualitative and verbose data using natural language processing (NLP) computational techniques to map anatomical phenotypic similarities using concept mapping to gene groups and pathways. These phenotype groups are can then be queried using descriptors generated by a screener to identify putative pathways being regulated by the small molecule of interest.
2) The identification of what biomolecule a small molecule interacts with to cause a phenotypic change is referred to as “target deconvolution”. Traditional modes of target deconvolution include affinity based approaches that are highly variable in their effectiveness and are incredibly labor intensive. This step is the transition from teratology to chemical biology, as such is also a step of value generation. To address this we are developing a tool based on classical forward genetics principles, that “like phenotypes will be in shared pathways”. This algorithm utilizes the wealth of mutant and knockdown data that is publically available for zebrafish as qualitative and verbose data using natural language processing (NLP) computational techniques to map anatomical phenotypic similarities using concept mapping to gene groups and pathways. These phenotype groups are can then be queried using descriptors generated by a screener to identify putative pathways being regulated by the small molecule of interest.

3) Once a small molecule and its molecular target are identified certain steps typically need to happen before these “Hit” compounds can be used as true in vivo probes. The most important of these steps involves medicinal chemistry and pharmacology. The criteria of a good in vivo probe are submicromolar potency, good solubility to assure delivery, and good biological stability and pharmacodynamics. These are properties are obtained through the process of structure-activity relationships(SAR). While traditional iterative medicinal chemistry approaches are used for proteins that have no "solved" structure, we utilize structure based computer-aided drug discovery techniques that include molecular modeling, docking, molecular dynamics and denovo small molecule design.
A Beating Heart Fueling Discovery
In addition to investigating morphological changes in development, the zebrafish also serves as an excellent model for cardiac physiology. The human heart beats an average of 60 beats per minute (bpm), the commonly used rodents models are 300-800 bpm. The fast rate of a rodent heart is due to fundamental differences in how the contraction-relaxation cycle is regulated. This difference has served as a stumbling block for early drug discovery as other mammalian models such as rabbit (120bpm) and dog (80 bpm), are both significantly larger and represent a significant cost increase which is not tenable for high risk "early discovery" programs. By comparison, a zebrafish heart rate is ~60bpm embryonically and adults are 120bpm. This comparable to human rate is accompanied by similar dynamics and mechanisms regulating the contraction-relaxation cycle of the heart. The two chambers of a zebrafish heart are easily seen and monitored without the need for highly sophisticated echocardiograms. Above is a video of a zebrafish heart beating.
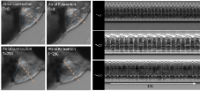
By using the zebrafish model we have identified novel modulators of cardiac activity. These modulators can slow down the heart or increase contractility. These molecules will help researchers identify new ways to modulate cardiac function and accelerate the discovery of novel therapies for heart disease which is the leading cause of mortality in the United States.
Next steps
This effort is supported by the NIH/NIGMS and we are sharing our compounds that have been discovered through the Chemical Phenomics Initiative. The chemical genetics component of our work is merely a starting point for collaborations using newly discovered tools and therapeutic development. Contact us for more information.
